Team: MYRIAD = « Modeling & analysis for medical imaging and Diagnosis »
Supervision: Nicolas DUCHATEAU (Associate Professor / U. Lyon 1), Patrick CLARYSSE (Senior Researcher / CNRS).
Context: The mechanisms of evolution of myocardial infarction and reperfusion injury are complex, and accompanied by shape and function abnormalities. Nevertheless, they are insufficiently characterized by clinical measures (lesion size and transmurality on MRI). We are developing statistical learning algorithms to better characterize these lesions and their evolution from several MRI acquisitions. In this context, having access to realistic biophysical models that simulate normal and pathological cardiac function is essential [CON-16] for the validation of these algorithms, but also to generate large realistic databases in support of real data for pre-training or data augmentation. Customization is an essential step for the realism of these simulations. The standard approach is to fit the model to an individual's data [LOP-19]; to generate large populations, the personalization is performed on all the individuals of a real population, giving a-posteriori a range of plausible model parameters. A synthetic population is thus obtained by running the simulations corresponding to values drawn in this interval. This approach is relevant for models, but not for models with a random component (e.g. to generate lesions of random location and shape [DUC-18]).
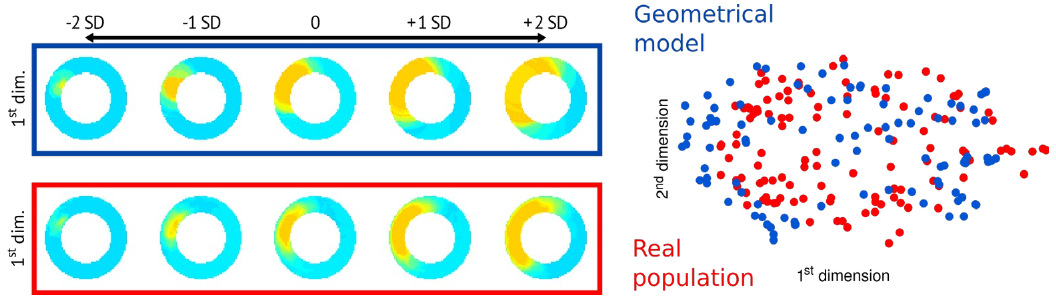
Figure: Currently achieved population-based personalization on 2D slices and a simple geometrical infarct model [MOM-21].
Objectives: We recently proposed an alternative population-wide personalization strategy [MOM-21]. It consists in finding the optimal parameters of the model to best match the virtual population with the real population studied. This strategy is partly comparable to the distributional matching of some learning methods (e.g. variational autoencoders), except that here the virtual population is generated from from the parameters of the customized model, and not from a latent space encoding the data.
We have demonstrated the potential of this strategy to customize simple geometrical models of myocardial infarction, in 2D, for a moderate size population. In the proposed project, we aim to:
- Demonstrate the viability of this 3D approach, on a broader population, which would allow the generation of more complex and realistic lesion shapes even with relatively simple geometrical models,
- Compare the relevance of different algorithms generating synthetic lesions after customization,
- Couple these results with MRI image simulation and/or biomechanical models to validate the learning methods we develop.
Practical information:
- The post-doc position is financed by the WP5 from Labex PRIMES of Université de Lyon, which focuses on simulation and modelling in medical imaging.
- It will take place at CREATIS (in the Doua campus in Villeurbanne), reference French lab in medical imaging, which consists of ~200 people grouped in 5 research teams. It will be supervised by N. Duchateau (Associate Professor) and P. Clarysse (Research Director), with strong interaction with clinical researchers from CHU St Etienne (P. Croisille – radiologist, and M. Viallon – medical physicist), and the team of PhD students and post-docs we supervise on related projects.
- Duration: 18 months, asap.
Profile: We look for a highly motivated post-doc candidate, with :
- Main background in applied mathematics and/or machine learning with strong interest for medical imaging applications,
- or main background in medical imaging with solid engineering skills in image processing and analysis,
- Good programming skills: Python (preferred). Complementary knowledge on Matlab and/or C++ may help.
- Fluent in English (reading, writing, speaking).
Contact: Send your CV, motivation letter, and academic record to: nicolas.duchateau@creatis.insa-lyon.fr
Bibliography:
[CON-16] Connolly & Bishop. Clin Med Insights Cardiol, 2016;10:27-40.
[DUC-18] Duchateau et al. IEEE T Med Imaging, 2018;37:755-66.
[LOP-19] Lopez-Perez et al. Front Physiol, 2019;10-580.
[MOM-21] Mom et al. Proc. FIMH, LNCS, 2021;12738:3-11.
