What is MicroVIP?
MicroVIP is a user friendly fluorescence microscopy image simulation and analysis tool deployed on Creatis' Virtual Imaging Platform (VIP). It enables simulation of 3D microscopy images issued from several microscopy techniques such as Wide-Field, Confocal, Structured Illumination Microscopy (SIM) and Light Sheet Microscopy (LSM).
It is meant to be used as a web application on VIP, but it is an open-source software and you can find its source code on Gitlab and its documentation on associated wiki.

Microscopy simulation: why?
Microscopy image simulation allows one to define and virtually perform microscopy experiments. It can be used to assess the effect of several parameters on the output microscopy images before conducting a real experiment. Moreover, this lets one generate a large number of images, together with their ground truth annotation, for an extremely low cost, making it a valuable tool for training or testing image analysis algorithms and validating their behavior when faced with several experimental conditions (sub-resolved or super-resolved images, variable cell phenotypes...). Many microscopy simulators have been proposed, but they usually are specific to a single microscopy technique. They also lack standardisation, and thus do not share common sets of inputs or outputs, common programmation languages, platforms, or types of interface. It is therfore a challenge for one to find the software they need with respect to their expertise, hardware constraints, experimental conditions, etc.
Why choosing MicroVIP?
With MicroVIP, we present a single user-friendly application gathering simulators for a wide range of microscopy techniques. In addition to offering a very easy-to-understand graphical interface, MicroVIP comes with comprehensive parameters customization capabilities for flexibility and adaptation to the experimenter needs. In addition to its predecessor's features, it notably allows simulation of the imaging process of cells in motion in a microfluidic system, with custom cell speed. Furthermore, MicroVIP implements a complete and self-sufficient simulation pipeline raher than only modeling the image acquisition process. It indeed integrates a module of generation of the ground truth cell chromatin chains and fluorescent markers positions, as well as another one performing commonly used features extraction methods on output simulated microscopy image.
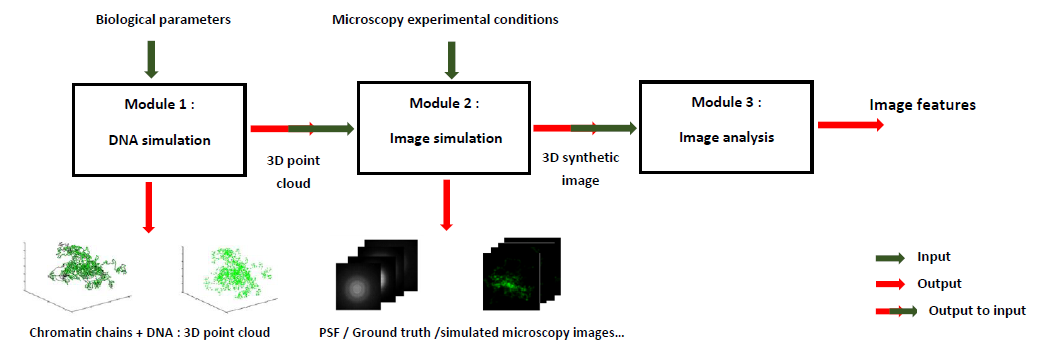
Where to find MicroVIP?
Project's context
MicroVIP has been developped at Creatis in the context of the three years (2018 - 2021) European project PROCHIP, carried out by an international consortium of 6 organizations from three European countries (Italy, France and England), led by the Italian National Reasearch Council. For more information, please visit PROCHIP official website.
Related publications:
AHMAD, Ali, FRINDEL, Carole, et ROUSSEAU, David. Detecting differences of fluorescent markers distribution in single cell microscopy: textural or pointillist feature space?. Frontiers in Robotics and AI, 2020, vol. 7, p. 39.
A. Ahmad, C. Frindel and D. Rousseau, "Sorting cells from fluorescent markers organization in confocal microscopy: 3D versus 2D images," 2020 Tenth International Conference on Image Processing Theory, Tools and Applications (IPTA), Paris, 2020, pp. 1-6, doi: 10.1109/IPTA50016.2020.9286463.
ALI, Ahmad, FRINDEL, Carole, et ROUSSEAU, David. Détection de différence de densité de marqueurs fluorescents en microscopie superrésolue: approche pointilliste ou texturale?. In : XXVIIe colloque Gretsi. 2019.
AHMAD, Ali, RASTI, Pejman, FRINDEL, Carole, et al. Deep learning based detection of cells in 3D light sheet fluorescence microscopy. In : Quantitative BioImaging Conference (QBI 2019). 2019.
T. Glatard, C. Lartizien, B. Gibaud, R. Ferreira da Silva, G. Forestier, F. Cervenansky, M. Alessandrini, H. Benoit-Cattin, O. Bernard, S. Camarasu-Pop, N. Cerezo, P. Clarysse, A. Gaignard, P. Hugonnard, H. Liebgott, S. Marache, A. Marion, J. Montagnat, J. Tabary, and D. Friboulet, A Virtual Imaging Platform for multi-modality medical image simulation, IEEE Transactions on Medical Imaging, vol. 32, no. 1, pp. 110-118, 2013


