Publications
In the field of forensic anthropology, researchers aim to identify anonymous human remains and determine the cause and circumstances of death from skeletonized human remains. Sex determination is a fundamental step of this procedure because it influences the estimation of other traits, such as age and stature. Pelvic bones are especially dimorphic, and are thus the most useful bones for sex identification. Sex estimation methods are usually based on morphologic traits, measurements, or landmarks on the bones. However, these methods are time-consuming and can be subject to inter- or intra-observer bias. Sex determination can be done using dry bones or CT scans. Recently, artificial neural networks (ANN) have attracted attention in forensic anthropology. Here we tested a fully automated and data-driven machine learning method for sex estimation using CT-scan reconstructions of coxal bones. We studied 580 CT scans of living individuals. Sex was predicted by two networks trained on an independent sample: a disentangled variational auto-encoder (DVAE) alone, and the same DVAE associated with another classifier (Crecon). The DVAE alone exhibited an accuracy of 97.9%, and the DVAE + Crecon showed an accuracy of 99.8%. Sensibility and precision were also high for both sexes. These results are better than those reported from previous studies. These data-driven algorithms are easy to implement, since the pre-processing step is also entirely automatic. Fully automated methods save time, as it only takes a few minutes to pre-process the images and predict sex, and does not require strong experience in forensic anthropology.

Facial expression generation is one of the most challenging and long-sought aspects of character animation, with many interesting applications. This challenging task, traditionally having relied heavily on digital craftspersons, remains yet to be explored. In this paper, we introduce a generative framework for generating 3D facial expression sequences (i.e. 4D faces) that can be conditioned on different inputs to animate an arbitrary 3D face mesh. It is composed of two tasks: (1) Learning the generative model that is trained over a set of 3D landmark sequences, and (2) Generating 3D mesh sequences of an input facial mesh driven by the generated landmark sequences. The generative model is based on a Denoising Diffusion Probabilistic Model (DDPM), which has achieved remarkable success in generative tasks of other domains. While it can be trained unconditionally, its reverse process can still be conditioned by various condition signals. This allows us to efficiently develop several downstream tasks involving various conditional generation, by using expression labels, text, partial sequences, or simply a facial geometry. To obtain the full mesh deformation, we then develop a landmark-guided encoder-decoder to apply the geometrical deformation embedded in landmarks on a given facial mesh. Experiments show that our model has learned to generate realistic, quality expressions solely from the dataset of relatively small size, improving over the state-of-the-art methods. Videos and qualitative comparisons with other methods can be found at https://github.com/ZOUKaifeng/4DFM.

Two recent works have shown the benefit of modeling both high-level factors and their related features to learn disentangled representations with variational autoencoders (VAE). We propose here a novel VAE-based approach that follows this principle. Inspired by conditional VAE, the features are no longer treated as random variables over which integration must be performed. Instead, they are deterministically computed from the input data using a neural network whose parameters can be estimated jointly with those of the decoder and of the encoder. Moreover, the quality of the generated images has been improved by using discrete latent variables and a two-step learning procedure, which makes it possible to increase the size of the latent space without altering the disentanglement properties of the model. Results obtained on two different datasets validate the proposed approach that achieves better performance than the two aforementioned works in terms of disentanglement, while providing higher quality images.

Herein, a multistep synthesis method was implemented to prepare nanoscale core@shell spherical heterostructures for H2 optical detection. The resulting nanoheterostructures are built with gold cores and silica shells further coated by a thin layer of spherical palladium nanoparticles. Samples with various thicknesses of silica and densities of palladium nanoparticles were synthesized and characterized by transmission electron microscopy. In particular, the three-dimensional core@shell configuration was confirmed by electron tomography. The optical properties of the obtained nanostructures were first analyzed by optical absorbance measurements, and their potential use for H2 detection was evaluated by simulations based on the discrete dipole approximation method. Thus, the optical response of the studied nanostructures is investigated theoretically when the Pd layer is hydrogenated, showing the great sensitivity achievable with these nanostructures.

Neural network-based classification methods are often criticized for their lack of interpretability and explainability. By highlighting the regions of the input image that contribute the most to the decision, saliency maps have become a popular method to make neural networks interpretable. In medical imaging, they are particularly well-suited for explaining neural networks in the context of abnormality localization. Nevertheless, they seem less suitable for classification problems in which the features that allow distinguishing classes are spatially correlated and scattered. We propose here a novel paradigm based on Disentangled Variational Auto-Encoders. Instead of seeking to understand what the neural network has learned or how prediction is done, we seek to reveal class differences. This is achieved by transforming the sample from a given class into the “same” sample but belonging to another class, thus paving the way to easier interpretation of class differences. Our experiments in the context of automatic sex determination from hip bones show that the obtained results are consistent with expert knowledge. Moreover, the proposed approach enables us to confirm or question the choice of the classifier or eventually to doubt it.

Most of previous semi-supervised methods that seek to obtain disentangled representations using variational autoencoders divide the latent representation into two components: the non-interpretable part and the disentangled part that explicitly models the factors of interest. With such models, any features associated with high-level factors are not explicitly modeled, so that they can either be lost, or at best entangled in the other latent variables, thus leading to bad disentanglement properties. To address this problem, we propose a novel conditional dependency structure where both the labels and their features belong to the latent space. We show using the CelebA dataset that the proposed model can learn meaningful representations, and we provide quantitative and qualitative comparisons with other approaches that show the effectiveness of the proposed method.

Purpose: We propose to learn a 3D keypoint descriptor which we use to match keypoints extracted from full-body CT scans. Our methods are inspired by 2D keypoint descriptor learning, which was shown to outperform hand-crafted descriptors. Adapting these to 3D images is challenging because of the lack of labelled training data and high memory requirements. Method : We generate semi-synthetic training data. For that, we first estimate the distribution of local affine inter-subject transformations using labelled anatomical landmarks on a small subset of the database. We then sample a large number of transformations and warp unlabelled CT scans, for which we can subsequently establish reliable keypoint correspondences using guided matching. These correspondences serve as training data for our descriptor, which we represent by a CNN and train using the triplet loss with online triplet mining. Results : We carry out experiments on a synthetic data reliability benchmark and a registration task involving 20 CT volumes with anatomical landmarks used for evaluation purposes. Our learned descriptor outperforms the 3D-SURF descriptor on both benchmarks while having a similar runtime. Conclusion: We propose a new method to generate semi-synthetic data and a new learned 3D keypoint descriptor. Experiments show improvement compared to a hand-crafted descriptor. This is promising as literature has shown that jointly learning a detector and a descriptor gives further performance boost.

Computational anatomy focuses on the analysis of the human anatomical vari- ability. Typical applications are the discovery of differences across healthy and sick subjects and the classification of anomalies. A fundamental tool in computa- tional anatomy, which forms the central focus of this paper, is the computation of point correspondences across volumes (3D images) such as Computed To- mography (CT) volumes, for multiple subjects. More specifically, we consider automatically detected keypoints and their local descriptors, computed from the image or volume patch surrounding each keypoint. Theses descriptors are essential because they must be discriminant and repeatable [5,10]. Learned de- scriptors based on Convolutional Neural Networks (CNN) have recently shown great success for 2D images [4]. However, while classical 2D image descriptors were extended to volumes [1], recent learning-based approaches have been lim- ited to 2D detection and description. The extension to 3D descriptors was only proposed in [6], in the context of image retrieval. We propose a methodology to construct these learned volume keypoint descriptors. The main difficulty is to define a sound training approach, combining a training dataset and a loss func- tion. In short, we propose to generate semi-synthetic data by transforming real volumes and to use a triplet loss inspired by 2D descriptor learning. Our exper- imental results show that our learned descriptor outperforms the hand-crafted descriptor 3D-SURF [1], a 3D extension of SURF, with similar runtime.

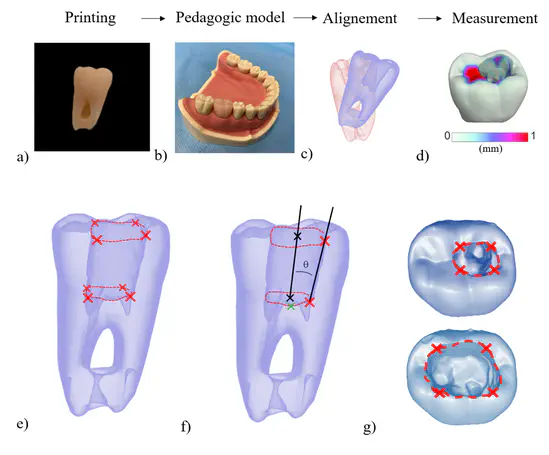
The instrumental fracture is a common endodontic complication that is treated by surgical or non-surgical removal approaches. However, no tool exists to help the clinician to choose between available strategies, and decision-making is mostly based on clinical judgment. Digital solutions, such as Finite Element Analysis (FEA) and Virtual Treatment Planning (VTP), were recently proposed in maxillofacial surgery. The aim of the current study is to present a digital tool to help decide between non-surgical and surgical strategies in a clinical situation of a fractured instrument. Five models have been created: the initial state of the patient, two non-surgical removal strategies using a low or high root canal enlargement, and two surgical removal strategies using a 3- or 6-mm apicoectomy. Results of the VTP found a risk of perforation for the non-surgical strategies and sinus proximity for surgical ones. FEA showed the lowest mechanical risk for the apicoectomy strategy. A 3-mm apicoectomy approach was finally chosen and performed. In conclusion, this digital approach could offer a promising decision support for instrument removal by planning the treatment and predicting the mechanical impact of each strategy, but further investigations are required to confirm its relevance in endodontic practice.

This paper presents an improvement of the keypoint transfer method for the segmentation of 3D medical images. Our approach is based on 3D SURF keypoint extraction, instead of 3D SIFT in the original algorithm. This yields a significantly higher number of keypoints, which allows to use a local segmentation transfer approach. The resulting segmentation accuracy is significantly increased, and smaller organs can be segmented correctly. We also propose a keypoint selection step which provides a good balance between speed and accuracy. We illustrate the efficiency of our approach with comparisons against state of the art methods.

In this work, we propose an experimental approach allowing for the identification and the subsequent quantification of nanoparticles crystallographic facets, based on 3D data obtained using Transmission Electron Microscopy (TEM). The particle shape and faceting can be determined using Electron Tomography (ET) combined with High-Resolution TEM (HR-TEM). The quantitative analysis of faceting is carried out on the particles 3D model using a geometrical approach that automatically detects planar regions on particle boundaries. In order to check the reliability of our approach, we analysed palladium particles confined inside or located at the outer surface of a mesoporous silica shell (Pd@SiO 2 ) after an annealing treatment at 250°C under H 2 for 12 h. From the materials perspective, the aim of the current work is to investigate the encapsulation effect of silica (SiO 2 ) shells on the change in morphology and faceting of palladium NPs during such annealing treatment.

We present a novel algorithm for Fast Registration Of image Groups (FROG), applied to large 3D image groups. Our approach extracts 3D SURF keypoints from images, computes matched pairs of keypoints and registers the group by minimizing pair distances in a hubless way i.e. without computing any central mean image. Using keypoints significantly reduces the problem complexity compared to voxel-based approaches, and enables us to provide an in-core global optimization, similar to the Bundle Adjustment for 3D reconstruction. As we aim to register images of different patients, the matching step yields many outliers. Then we propose a new EM-weighting algorithm which efficiently discards outliers. Global optimization is carried out with a fast gradient descent algorithm. This allows our approach to robustly register large datasets. The result is a set of diffeomorphic half transforms which link the volumes together and can be subsequently exploited for computational anatomy and landmark detection. We show experimental results on whole-body CT scans, with groups of up to 103 volumes. On a benchmark based on anatomical landmarks, our algorithm compares favorably with the star-groupwise voxel-based ANTs and NiftyReg approaches while being much faster. We also discuss the limitations of our approach for lower resolution images such as brain MRI.

Ce mémoire se découpe en 4 parties. La première partie chapitre aborde la compression de données 3D, et particulièrement la transmission progressive de maillages surfaciques triangulaires d’objets 3D, et par exmple, le Raffinement Incrémental Paramétrique, y sera détaillé. La deuxième partie concerne mes contributions dans le domaine de la génération de maillages. Une de mes motivations principales dans ce contexte est la proposition d’algorithmes robustes et rapides, capables de s’adapter à une multitude de données en entrée. Une contribution importante est la proposition d’une approche basée sur le partitionnement d’ensembles discrets approximant un diagramme de Voronoï centroïdal. La troisième partie concerne la segmentation et le recalage d’images médicales 3D, proposant à la fois des approches interactives et automatiques. La nature très hétérogène des images (modalités d’imagerie différentes, parties du corps imagées différentes) rend le problème générique très difficile à résoudre. La dernière partie présente une synthèse des perspectives de recherche, avec une orientation marquée sur l’analyse à grande échelle d’images médicales 3D

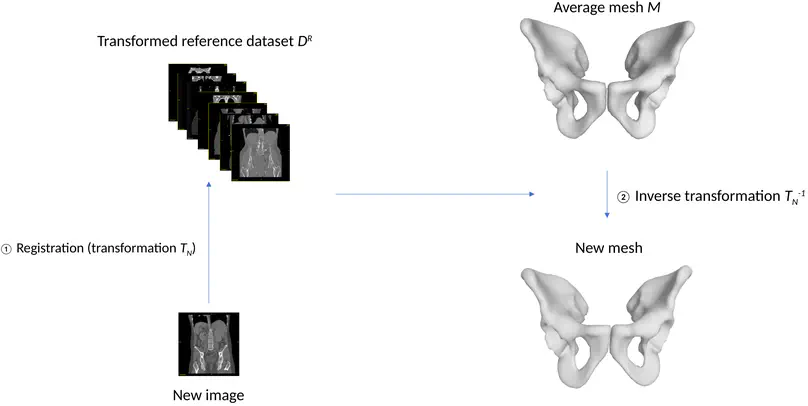
The purpose of this study is to assess the relevance of computational anatomy for the sex determination in forensic anthropology. A novel groupwise registration algorithm is used, based on keypoint extraction, able to register several hundred full body images in a common space. Experiments were conducted on 83 CT scanners of living individuals from the public VISCERAL database. In our experiments, we first verified that the well-known criteria for sex discrimination on the hip-bone were well preserved in mean images. In a second experiment, we have tested semi-automatic positioning of anatomical landmarks to measure the relevance of groupwise registration for future research. We applied the Probabilistic Sex Diagnosis tool on the predicted landmarks. This resulted in 62% of correct sex determinations, 37% of undetermined cases, and 1% of errors. The main limiting factors are the population sample size and the lack of precision for the initial manual positioning of the landmarks in the mean image. We also give insights on future works for robust and fully automatic sex determination.

We propose an automatic multiorgan segmentation method for 3D radiological images of different anatomical content and modality. The approach is based on a simultaneous multilabel Graph Cut optimization of location, appearance and spatial configuration criteria of target structures. Organ location is defined by target-specific probabilistic atlases (PA) constructed from a training dataset using a fast (2+1)D SURF-based multiscale registration method involving a simple 4-parameter transformation. PAs are also used to derive target-specific organ appearance models represented as intensity histograms. The spatial configuration prior is derived from shortest-path constraints defined on the adjacency graph of structures. Thorough evaluations on Visceral project benchmarks and training dataset, as well as comparisons with the state of the art confirm that our approach is comparable to and often outperforms similar approaches in multiorgan segmentation, thus proving that the combination of multiple suboptimal but complementary information sources can yield very good performance.

The thermal stability of core-shell Pd@SiO2 nanostructures was for the first time directly monitored in the real space by using the in-situ Environmental Transmission Electron Microscopy (E-TEM) at atmospheric pressure coupled with Electron Tomography (ET) realized on the same particles. The core Palladium (Pd) particles, with octahedral or icosahedral shapes, were followed by E-TEM during thermal heating in an H2 and air flow at atmospheric pressure. In a first step, their morphology/faceting evolution was investigated under H2 gas up to 400 °C by E-TEM combined with ET experiments performed on the same particles before and after the in situ treatment. As a result, we observed the formation of new Pd particles inside the silica shell due to the thermally activated diffusion from the core particle, as well as the dependence of the shape and faceting thermal evolution on the initial structure of the particles: the octahedral monocrystalline NPs were found to be less stable than the icosahedral ones, the atom diffusion leads to a decrease of the particles size due to the high atom migration towards the silica shell; the icosahedral polycrystalline NPs do not exhibit morphology/faceting change, the atom diffusion within the particle is favoured against diffusion towards the silica shell, in agreement with a high amount of crystallographic defects. In the second part, the Pd@SiO2 NPs behaviour at high temperatures (up to 1000 °C) was investigated by E-TEM under reductive or oxidative conditions and found to be dependant from the thermal evolution of the silica shell: (1) under H2, the silica is densified and lost its porous structure, the Pd cores remaining thus encapsulated; (2) under air, the silica porosity is maintained and the increase of the temperature leads to an enhancement of the diffusion mechanism from the core towards the external surface of the silica, at 850 °C the totality of the Pd core being thus expulsed outside the silica shell.

We propose a new shape analysis approach based on the non-local analysis of local shape variations. Our method relies on a novel description of shape variations, called Local Probing Field (LPF), which describes how a local probing operator transforms a pattern onto the shape. By carefully optimizing the position and orientation of each descriptor, we are able to capture shape similarities and gather them into a geometrically relevant dictionary over which the shape decomposes sparsely. This new representation permits to handle shapes with mixed intrinsic dimensionality (e.g. shapes containing both surfaces and curves) and to encode various shape features such as boundaries. Our shape representation has several potential applications; here we demonstrate its efficiency for shape resampling and point set denoising for both synthetic and real data.

We propose a method for the automatic positioning of pre-defined landmarks on 3-D models of anatomical structures. We exploit a group of atlases consisting of multiple triangular meshes for which the defined landmarks have been placed by experts. We compute an initial coarse global registration of the patient mesh with an expert mesh by using a curvature-enhanced Iterative Closest Point (ICP) algorithm. Adaptive local rigid registrations refine the fit for the projection of reference landmarks onto the surface of the patient structure. An automatic selection based on a fit criterion computes a final position for each landmark. Our positioning method improves the efficiency of the positioning task, being completely unsupervised and yielding results competitive with those of the manual positioning. We provide comparisons with previous works of the literature. The automatic positioning for each target structure is completely reproducible as opposed to manual positioning, affected by intra-operator variability.

We propose a generic method for the automatic multiple-organ segmentation of 3D images based on a multilabel graph cut optimization approach which uses location likelihood of organs and prior information of spatial relationships between them. The latter is derived from shortest-path constraints defined on the adjacency graph of structures and the former is defined by probabilistic atlases learned from a training dataset. Organ atlases are mapped to the image by a fast (2+1)D hierarchical registration method based on SURF keypoints. Registered atlases are also used to derive organ intensity likelihoods. Prior and likelihood models are then introduced in a joint centroidal Voronoi image clustering and graph cut multiobject segmentation framework. Qualitative and quantitative evaluation has been performed on contrast-enhanced CT and MR images from the VISCERAL dataset.

We propose a hubless medical image registration scheme, able to conjointly register massive amounts of images. Exploiting 3D points of interest combined with global optimization, our algorithm allows partial matches, does not need any prior information (full body image as a central patient model) and exhibits very good robustness by exploiting inter-volume relationships. We show the efficiency of our approach with the rigid registration of 400 CT volumes, and we provide an eye-detection application as a first step to patient image anonymization.

Real-time 3D Echocardiography (RT3DE) has been proven to be an accurate tool for left ventricular (LV) volume assessment. However, identification of the LV endocardium remains a challenging task, mainly because of the low tissue/blood contrast of the images combined with typical artifacts. Several semi and fully automatic algorithms have been proposed for segmenting the endocardium in RT3DE data in order to extract relevant clinical indices, but a systematic and fair comparison between such methods has so far been impossible due to the lack of a publicly available common database. Here, we introduce a standardized evaluation framework to reliably evaluate and compare the performance of the algorithms developed to segment the LV border in RT3DE. A database consisting of 45 multivendor cardiac ultrasound recordings acquired at different centers with corresponding reference measurements from 3 experts are made available. The algorithms from nine research groups were quantitatively evaluated and compared using the proposed online platform. The results showed that the best methods produce promising results with respect to the experts’ measurements for the extraction of clinical indices, and that they offer good segmentation precision in terms of mean distance error in the context of the experts’ variability range. The platform remains open for new submissions.

Most surfaces, be it from a fine-art artifact or a mechanical object, are characterized by a strong self-similarity. This property finds its source in the natural structures of objects but also in the fabrication processes: regularity of the sculpting technique, or machine tool. In this paper, we propose to exploit the self-similarity of the underlying shapes for compressing point cloud surfaces which can contain millions of points at a very high precision. Our approach locally resamples the point cloud in order to highlight the self-similarity of the shape, while remaining consistent with the original shape and the scanner precision. It then uses this self-similarity to create an ad hoc dictionary on which the local neighborhoods will be sparsely represented, thus allowing for a light-weight representation of the total surface. We demonstrate the validity of our approach on several point clouds from fine- arts and mechanical objects, as well as a urban scene. In addition, we show that our approach also achieves a filtering of noise whose magnitude is smaller than the scanner precision.

Magnetic resonance imaging (MRI) based 3D reconstructions were used to derive accurate quantitative data on body volume and functional skin surface areas involved in water transfer in the Palmate Newt (Lissotriton helveticus (Razoumovsky, 1789)). Body surface area can be functionally divided into evaporative surface area that interacts with the atmosphere and controls the transepidermal evaporative water loss (TEWL); ventral surface area in contact with the substratum that controls transepidermal water absorption (TWA); and skin surface area in contact with other skin surfaces when amphibians adopt water-conserving postures. We generated 3D geometries of the newts via volume-rendering by a “segmentation” process carried out using a graph-cuts algorithm and a Web-based interface. The geometries reproduced the two postures adopted by the newts, i.e., an I-shaped posture characterized by a straight body without tail coiling and an S-shaped posture where the body is huddled up with the tail coiling along it. As a guide to the quality of the surface area estimations, we compared measurements of TEWL rates between living newts and their agar replicas (reproducing their two postures) at 20°C and 60% relative humidity. Whereas the newts did not show any physiological adaptations to restrain evaporation, they expressed an efficient S-shaped posture with a resulting water economy of 22.9%, which is very close to the 23.6% reduction in evaporative surface area measured using 3D analysis.

We derive shortest-path constraints from graph models of structure adjacency relations and introduce them in a joint centroidal Voronoi image clustering and Graph Cut multiobject semiautomatic segmentation framework. The vicinity prior model thus defined is a piecewise-constant model incurring multiple levels of penalization capturing the spatial configuration of structures in multiobject segmentation. Qualitative and quantitative analysis and comparison with a Potts prior-based approach and our previous contribution on synthetic, simulated and real medical images show that the vicinity prior allows for the correct segmentation of distinct structures having identical intensity profiles and improves the precision of segmentation boundary placement while being fairly robust to clustering resolution. The clustering approach we take to simplify images prior to segmentation strikes a good balance between boundary adaptivity and cluster compactness criteria furthermore allowing to control the trade-off. Compared to a direct application of segmentation on voxels, the clustering step improves the overall runtime and memory footprint of the segmentation process up to an order of magnitude without compromising the quality of the result.

We propose a web-accessible image visualization and processing framework well-suited for medical applications. Exploiting client-side HTML5 and WebGL technologies, our proposal allows the end-user to efficiently browse and visualize volumic images in an Out-Of-Core (OOC) manner, annotate and apply server-side image processing algorithms and interactively visualize 3D medical models. Server-side implementation is driven by a file-based, simple, robust and flexible Remote Procedure Call (RPC) scheme well suited for heterogeneous applications. We demonstrate the efficiency of our approach with both an interactive medical image segmentation and a 3D rendering of segmented anatomical structures. As a secondary contribution, we improve the segmentation algorithm with the introduction of user-defined anatomical priors.

Trabecular bone is made of a complex network of plate and rod structures, the proportion of which evolves with age or disease. Thus the identification of trabecular plates and rods is important in understanding bone fragility. We propose a novel approach based on 3D multi-scale adjacency graph analysis of high resolution 3D tomographic images of bone structures. The purpose of this new method is to classify each voxel of the 3D images in two classes : plate and rod voxels. We show that the use of a multi-scale framework is very efficient at detecting rods with different sizes. We present applications of our method to both synthetic images and experimental bone synchrotron radiation micro-CT images.

In this paper, we present a wavelet-based progressive compression method for isomorphic 3-D mesh sequence with constant connectivity. Our method reduces the spatial and temporal redundancy by using both spatial and temporal wavelet analysis. To encode geometry information, each mesh frame is decomposed into a base mesh and its spatial wavelet coefficients of each resolution level by spatial wavelet analysis filter bank. The spatially transformed sequence is decomposed into several sub-band signals by temporal wavelet analysis filter bank. The resulting signal is encoded by using an arithmetic coder. Since an isomorphic mesh sequence has the same connectivity over all frames, the connectivity information is encoded only for the first mesh frame. The proposed method enables both progressive representation and lossless compression in a single framework by multi-resolution wavelet analysis with a perfect reconstruction filter bank. Our method is compared with several conventional techniques including our previous work.

In this paper, we propose a novel progressive lossless mesh compression algorithm based on Incremental Parametric Refinement, where the connectivity is uncontrolled in a first step, yielding visually pleasing meshes at each resolution level while saving connectivity information compared to previous approaches. The algorithm starts with a coarse version of the original mesh, which is further refined by means of a novel refinement scheme. The mesh refinement is driven by a geometric criterion, in spirit with surface reconstruction algorithms, aiming at generating uniform meshes. The vertices coordinates are also quantized and transmitted in a progressive way, following a geometric criterion, efficiently allocating the bit budget. With this assumption, the generated intermediate meshes tend to exhibit a uniform sampling. The potential discrepancy between the resulting connectivity and the original one is corrected at the end of the algorithm. We provide a proof-of-concept implementation, yielding very competitive results compared to previous works in terms of rate/distortion trade-off.

In this paper, we propose a new algorithm to mesh implicit surfaces which produces meshes both with a good triangle aspect ratio as well as a good approximation quality. The number of vertices of the output mesh is defined by the end-user. For this goal, we perform a two-stage processing : an initialization step followed by an iterative optimization step. The initialization step consists in capturing the surface topology and allocating the vertex budget. The optimization algorithm is based on a variational vertices relaxation and triangulation update. In addition a gradation parameter can be defined to adapt the mesh sampling to the curvature of the implicit surface. We demonstrate the efficiency of the approach on synthetic models as well as real-world acquired data, and provide comparisons with previous approaches.

In this paper, we propose a novel tetrahedral mesh generation algorithm, which takes volumic data (voxels) as an input. Our algorithm performs a clustering of the original voxels within a variational framework. A vertex replaces each cluster and the set of created vertices is triangulated in order to obtain a tetrahedral mesh, taking into account both the accuracy of the representation and the elements quality. The resulting meshes exhibit good elements quality with respect to minimal dihedral angle and tetrahedra form factor. Experimental results show that the generated meshes are well suited for Finite Element Simulations.

In this paper, we present a novel method for medial axis approximation based on Constrained Centroidal Voronoi Diagram of discrete data (image, volume). The proposed approach is based on the shape boundary subsampling by a clustering approach which generates a Voronoi Diagram well suited for Medial Axis extraction. The resulting Voronoi Diagram is further filtered so as to capture the correct topology of the medial axis. The resulting medial axis appears largely invariant with respect to typical noise conditions in the discrete data. The method is tested on various synthetic as well as real images. We also show an application of the approximate medial axis to the sizing field for triangular and tetrahedral meshing

In this paper, we propose a generic framework for 3D surface remeshing. Based on a metric-driven Discrete Voronoi Diagram construction, our output is an optimized 3D triangular mesh with a user defined vertex budget. Our approach can deal with a wide range of applications, from high quality mesh generation to shape approximation. By using appropriate metric constraints the method generates isotropic or anisotropic elements. Based on point-sampling, our algorithm combines the robustness and theoretical strength of Delaunay criteria with the efficiency of entirely discrete geometry processing . Besides the general described framework, we show experimental results using isotropic, quadric-enhanced isotropic and anisotropic metrics which prove the efficiency of our method on large meshes, for a low computational cost.

In this paper, we propose an adaptive polygonal mesh coarsening algorithm. This approach is based on the clustering of the input mesh triangles, driven by a discretized variationnal definition of centroidal tesselations. It is able to simplify meshes with high complexity i.e. meshes with a large number of vertices and high genus. We demonstrate the ability our scheme to simplify meshes according to local features such as curvature measures. We also introduce an initial sampling strategy which speeds up the algorithm, an on-the-fly checking step to guarantee the validity of the clustering, and a postprocessing step to enhance the quality of the approximating mesh. Experimental results show the efficiency of our scheme both in terms of speed and visual quality.

This paper introduces a novel partitioning algorithm for 3D polygonal meshes. The proposed approach is based on protrusion conquest which, for a given model, takes into account both the computed protrusion and the connectivity. The only constraint on the input mesh is that it must consist of one connected component. Our algorithm provides a good way to decompose the mesh into preceptually significant parts. The parts are further modeled by ellipsoids and a connectivity graph between them. This semantic representation is compliant to the perceptual shape description defined by the emergent standard MPEG-7

A new content based mesh design algorithm for video objects segmentation and tracking is proposed. The algorithm moves the mesh nodes along motion or luminance discontinuities in order to fit the objects boundaries. The motion inside adjacent triangular cells is estimated by competitive forward-backward matching. This approach allows the displacements of the nodes without the remeshing of occlusion regions : uncovered background nor background to be covered. The mesh is deformed under the minimization of an objective function. This function includes a motion energy, a spatial boundary energy and a temporal regularization energy. The motion energy is effective in textured regions while the boundary energy mainly operates in homogeneous one according to a boundary distance map. The temporal regularization energy keeps the objects tracking stable along the sequence due to its object-based approach. Experimental results with both synthetic and real world sequences prove the efficiency of the proposed approach and the cooperative action of the three energy terms.

This paper extends Lounsbery s multiresolution analysis wavelet-based theory for triangular 3D meshes, which can only be applied to regularly subdivided meshes and thus involves a remeshing of the existing 3D data. Based on a new irregular subdivision scheme, the proposed algorithm can be applied directly to irregular meshes, which can be very interesting when one wants to keep the connectivity and geometry of the processed mesh completely unchanged. This is very convenient in CAD (Computer-Assisted Design), when the mesh has attributes such as texture and color information, or when the 3D mesh is used for simulations, and where a different connectivity could lead to simulation errors. The algorithm faces an inverse problem for which a solution is proposed. For each level of resolution, the simplification is processed in order to keep the mesh as regular as possible. In addition, a geometric criterion is used to keep the geometry of the approximations as close as possible to the original mesh. Several examples on various reference meshes are shown to prove the efficiency of our proposal.

We present a novel clustering algorithm for polygonal meshes which approximates a Centroidal Voronoi Diagram construction. The clustering provides an efficient way to construct uniform tessellations, and therefore leads to uniform coarsening of polygonal meshes, when the output triangulation has many fewer elements than the input mesh. The mesh topology is also simplified by the clustering algorithm. Based on a mathematical framework, our algorithm is easy to implement, and has low memory requirements. We demonstrate the efficiency of the proposed scheme by processing several reference meshes having up to 1 million triangles and very high genus within a few minutes on a low-end computer.

This paper proposes a new lossy to lossless progressive compression scheme for triangular meshes, based on a wavelet multiresolution theory for irregular 3D meshes. Although remeshing techniques obtain better compression ratios for geometric compression, this approach can be very effective when one wants to keep the connectivity and geometry of the processed mesh completely unchanged. The simplification is based on the solving of an inverse problem. Optimization of both the connectivity and geometry of the processed mesh improves the approximation quality and the compression ratio of the scheme at each resolution level. We show why this algorithm provides an efficient means of compression for both connectivity and geometry of 3D meshes and it is illustrated by experimental results on various sets of reference meshes, where our algorithm performs better than previously published approaches for both lossless and progressive compression.

We propose a new subdivision scheme derived from the Lounsbery’s 1:4 face split, allowing multiresolution analysis of irregularly subdivided triangular meshes by the wavelet transforms. Some experimental results on real medical meshes prove the efficiency of this approach in multiresolution schemes. In addition we show the effectiveness of the proposed algorithm for lossless compression.