Category: Image Processing
-
DLMI 2024
The 5th edtion of the school on Deep Learning for Medical Imaging (DLMI) held in Montréal from 8 to 12 of July and the asynchronous version one week after. More information about materials, speakers, hands-on : https://event.fourwaves.com/dlmi2024
-
CLANU 2021
Ressources pour le projet CLANU partie informatique pour la classification d’images d’IRM provennant de 3 séquences et coupées suivant 3 plans (problème de 9 classes) Enoncé de la partie informatique et code source C++
-
4th Edition of the Deep Learning Medical Imaging School
DLMI23: Fourth edition of the Deep learning for medical imaging school – Lyon April 17-21 2023 https://deepimaging2023.sciencesconf.org/ DeepImaging 2023 is organized by the LabEx Primes, Creatis, LabHC laboratories, the University of Sherbrooke and the ETS of Montreal. This school is intended for medical imaging beginners and experts (students, post-docs, research professionals, and professors) who are…
-
3rd edition of Deep Learning Medical Image School 2022
The third edition of the school is almost done. Thanks to participants and the Montreal organizing committee! We had an exciting week. The next edition (4th) will take place in Lyon. Dates are coming soon. Link to 3rd edition ressources: https://event.fourwaves.com/dlmi2022/pages
-
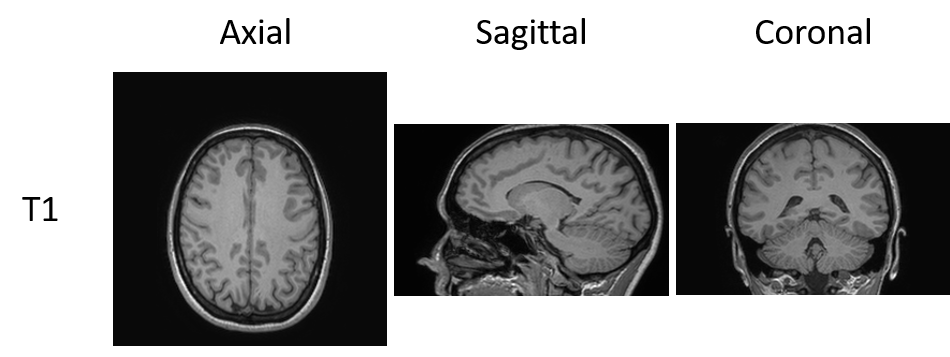
Machine learning medical image classification notebooks
Image classification is an interesting challenge that requires many images for training. Here we propose to explore 4 different machine learning classification approaches (from random forest to ResNet) on an image set created from the IXI data set using famous python packages: scikit-learn and Keras/TensorFlow. This example is split into 4 notebooks. The first notebook…
-
Practice on medical image segmentation with UNet
Here are the hands-on materials to practice deep learning segmentation with UNet (Keras/TF2): TP_UNET_TF2.zip This archive contains jupyter notebooks and necessary functions (i.e. all the code in python). See another post to install TF2/Keras with conda. The data are available here: dlss21_ho4_data.tar.gz The command line to extract the data is: $ tar xzf dlss21_ho4_data.tar.gz The…
-
Using Jupyter Lab and Tensorflow within a conda environment
1- before launching jupyter, check the environment kernel $ jupyter kernelspec list 2- if nothing appears looking “TF2.6”, add the kernel to jupyter by: $ ipython kernel install –name “TF2.6” –user 3- now you should see TF2.6 with the command: $ jupyter kernelspec list 4 -you can repeat the above kernel installation for other kernels…
-
Simple Filtering and Segmentation of medical image
Here is the notebook to start with SimpleITK and basics such as N4 bias field correction on MRI and segmentation using region growing and python programming. We also study the k-means segmentation. Python notebook : (zip)
-
Medical Deep Imaging spring school 2021
Our third edition of the spring/summer school on deep learning for medical images will be ‘virtual’ and from 19 to 24 of April 2021. Visit the official website here. As the previous edition, there are some lectures, practices, and social events. Most of the content will be available after the school. The first edition web…
-
Install Tensorflow2 or PyTorch/MONAI-dev with conda
Create a TensorFlow conda environment A short summary of conda utilization: This procedure is the same for PyTorch, just download monai-dev.yml and adapt the previous lines. This file includes MONAI-dev prerequisits. then finalize the MONAI-dev installation by first activate your env and then installing the MOANI-dev using (more information here):